Pathogen Evolutionary Genomics and Experimental Genetics
High adaptability to the microenvironment of the human host is an absolute requirement of all microbes that cause human diseases. Successful pathogens such as, Plasmodium falciparum, Mycobacterium tuberculosis, HIV-1 or SARS-CoV-2, while very different in complexity, share the capacity to acquire resistance, avoid elimination by the immune system, replicate or persist within the host and transmit.
The evolutionary and adaptive mechanisms that potentiate their capacity to thrive in the hostile environment of the human host are also involved in the response to antimicrobial treatment expediting the emerge of resistance. These processes are tightly regulated at the level of the genome and epigenome of the pathogens following fundamental molecular biology principals. Understanding the evolution of human pathogens and the use of computation systems biology approaches based on ‘OMICS’ technologies and bioinformatics allows staying ahead of the curve regarding emerging and re-emerging threats posed by infectious disease with emphasis on the challenge of antimicrobial resistance.
Also, applying functional and experimental genetics, that includes the technological breakthroughs of genome editing and modulation of gene expression at the RNA level, the team focus on understanding how pathogen genetic variations impact biological function leading to a functional validation of relevant molecular targets and biomarkers. Most importantly, the seamless integration between evolutionary genomics and experimental genetics has the potential to provide comprehensive novel insights into microbial mechanisms of pathogenicity that might open new avenues toward the design of novel, specific and more effective diagnostic applications, and therapeutics interventions.
Team Members


Nuno S. Osório

Ana Santos-Pereira

Claudia Fançony

Vera Cristina Aguieiras Triunfante

Carlos Magalhães

Flávia Figueira

Maria Pereira

Matheus Tavares

Pedro Araújo

Ruth Victoria Esho

Talita Nicolau

Vitória da Cunha Baptista

Beatriz Velosa da Fonseca

Bruno José Ferreira Freitas

Claudia Alejandra Cabrera Federo

Margarida Gonçalves
Sonia Carvalho
Team Members

Maria Isabel Veiga
Principal Investigator
Team coordinator

Nuno S. Osório
Senior Researcher

Ana Santos-Pereira
Postdoctoral Researcher

Claudia Fançony
Postdoctoral Researcher

Vera Cristina Aguieiras Triunfante
Affiliated Researcher

Carlos Magalhães
PhD Student

Maria Pereira
PhD Student

Pedro Araújo
PhD Student

Vitória da Cunha Baptista
PhD Student

Beatriz Velosa da Fonseca
MSc Student

Bruno José Ferreira Freitas
MSc Student

Claudia Alejandra Cabrera Federo
MSc Student

Filipa Tavares
MSc Student

Inês Gonçalves Geraldes
MSc Student

Mariagiulia Conte
MSc Student

Ruth Victoria Esho
Affiliated Researcher
Projects
- Ana Santos-Pereira
- There is still no cure for HIV infection and the extensive viral genetic diversity represents the one of the biggest obstacles to develop an effective therapy…
- Maria Isabel Veiga
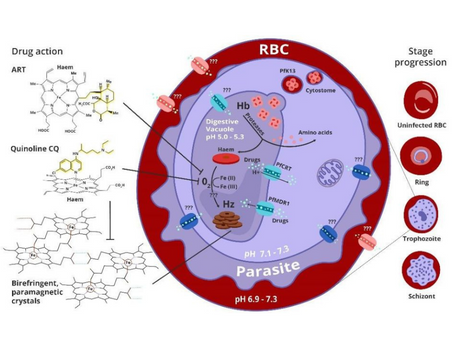
- The clinical manifestation of malaria starts when parasites invade human red blood cells (RBCs) and feed on haemoglobin, releasing a toxic by-product, polymerized by the parasite into the inert crystal hemozoin..
- Maria Isabel Veiga
- Hampering malaria control and elimination efforts is the resilient capacity of the parasite Plasmodium falciparum to develop resistance, including resistance to the presently…
- Nuno S. Osório
- Tuberculosis (TB) is one of the oldest and deadliest human infectious disease. Only innovative tools will allow realistic approximations to TB eradication…
- Nuno S. Osório
- The extremely high human-to-human transmissibility of SARS-CoV-2 allows the generation of thousands of mutations within its genome fueling the evolution and selection of several viral lineages with different characteristics…
- Claudia Fançony
- There is still no cure for HIV infection and the extensive viral genetic diversity represents the one of the biggest obstacles to develop an effective therapy…
Success Story
Selected Research Outputs
WO2021191882 ANTIMALARIAL AGENT, METHODS AND USES THEREOF (wipo.int)
WO2022070084 AUTOMATIC DEVICE FOR NON-INVASIVE MALARIA DIAGNOSIS THROUGH OPTICAL REFLECTANCE TECHNIQUES, METHODS AND USES THEREOF (wipo.int)
Frontiers | Plasmodium falciparum Drug Resistance Genes pfmdr1 and pfcrt In Vivo Co-Expression During Artemether-Lumefantrine Therapy (frontiersin.org)
The Future in Sensing Technologies for Malaria Surveillance: A Review of Hemozoin-Based Diagnosis | ACS Sensors
PEvoGen team main achievements:
– Contributed to the knowledge of the mechanisms of resistance to antimalarial treatments in the malaria parasite Plasmodium falciparum
– Improved antimalarial drug discovery by identifying new, essential, druggable targets and test compounds that may be advanced into drug candidates suitable for preclinical development and subsequent clinical testing in humans.
– Explored better malaria diagnosis, using differential physical properties of the malaria parasite regarding the hemozoin biological pathway.
– Highlighted relevant factors contributing to the colonization of human populations by viruses of different HIV-1 subtypes.
– Endeavored the challenge of investigating Mycobacterium tuberculosis mutations accumulated during the transmission between individuals infected in the same transmission chain.
– Advanced the study of phylogeny, transmission and dissemination of HIV-1, M. tuberculosis and more recently SARS-CoV-2 through Bioinformatics.
Former lab members
• Carla Sofia Calçada, PhD (Scientific Researcher, Toronto, Canada)
• Deisy Rocha, PhD (Postdoctoral Researcher, IPBS-Toulouse, France)
• Diana Granja, MSc (Bioengineer, Anocca biotech company, Sweden)
• Francisco Araujo, MD, MSc (Project leader, Autonomous University of Santo Domingo, Dominican Republic)
• Geniss Nhapulo, MD
• Isaac Sanchéz, MSc (Technical Coordinator at the National Reference Public Health Laboratory, Dominican Republic)
• Leyre Lau (PhD student, Karolinska Institute, Sweden)
• Miguel João Ferreira da Silva, PhD (Postdoctoral Researcher, Masaryk University, Czechia)
Evolutionary dynamics of HIV-1 subtype C in Brazil | Scientific Reports (nature.com)
Biomedicines | Free Full-Text | OmniSARS2: A Highly Sensitive and Specific RT-qPCR-Based COVID-19 Diagnostic Method Designed to Withstand SARS-CoV-2 Lineage Evolution (mdpi.com)
Plasmodium falciparum transcription in different clinical presentations of malaria associates with circulation time of infected erythrocytes | Nature Communications
Implication of SARS-CoV-2 evolution in the sensitivity of RT-qPCR diagnostic assays - The Lancet Infectious Diseases
Expansion of a Specific Plasmodium falciparum PfMDR1 Haplotype in Southeast Asia with Increased Substrate Transport | mBio
Mycobacterium tuberculosis associated with severe tuberculosis evades cytosolic surveillance systems and modulates IL-1β production | Nature Communications



Contact us
Phone: +351 253 604 967
Fax: +351 253 604 809
Email: icvs.sec@med.uminho.pt
Address
Life and Health Sciences
Research Institute (ICVS)
School of Medicine,
University of Minho,
Campus de Gualtar
4710-057 Braga
Portugal

Copyright ©2022 ICVS. All Rights Reserved



Copyright ©2022 ICVS. All Rights Reserved
Address
Life and Health Sciences
Research Institute (ICVS)
School of Medicine,
University of Minho,
Campus de Gualtar
4710-057 Braga
Portugal



Copyright ©2022 ICVS. All Rights Reserved
Address
Life and Health Sciences
Research Institute (ICVS)
School of Medicine,
University of Minho,
Campus de Gualtar
4710-057 Braga
Portugal